Site pages
Current course
Participants
General
Module 1. Phase Rule
Module 2. Fuels
Module 3. Colloids Classification, properties
Module 4. Corrosion Causes, type and methods of p...
Module 5. Water Hardness
Module 6. Scale and sludge formation in boilers, b...
Module 7. Analytical methods like thermo gravimetr...
Module 8. Nuclear radiation, detectors and analyti...
Module 9. Enzymes and their use in manufacturing o...
Module 10. Principles of Food Chemistry
Module 11. Lubricants properties, mechanism, class...
Module 12. Polymers type of polymerization, proper...
Lesson 19 Enzymes-II
19.1 Introduction
In this lesson, we will study mechanism of enzyme action, enzyme kinetics, Michaelis-Menten equation and regulation of enzyme activity.
19.2 Mechanism of Enzyme activity
All enzymes contain at least one active site (a specific region of enzyme) which combines with substrate (reacting molecules). The binding of substrate and enzyme causes change in shape of substrate molecules which leads to formation of new bonds or breakage of old bonds which leads to formation of product molecules. The products are released from enzyme surface and the enzyme molecules can enter other reaction cycle. In this way a small number of enzymes molecules change a large number of reactant molecules to form products.
It is found that enzymes are specific towards substrates. This can be explained on the basis of lock and key model. Consider the enzyme molecule to be a ‘lock’ and substrate molecules its ‘key’. We know that every lock and key combination is specific i.e. A lock can be opened only by a particular key. Similarly enzyme-substrate combination is specific. This happens because the shape of the active site of the enzyme and substrate are complimentary to each other similar to fitting of the puzzle pieces to form a complete picture as illustrated in figure no.19.1. This means that an enzyme molecule reacts with only one or very few similar compounds.
If a substrate molecule does not fit correctly to an enzyme, no reaction will take place as illustrated in Fig no.19.2
Figure 19.1 Figure showing combination of enzyme with correct substrate
Figure 19.2 Figure showing combination of enzyme with incorrect substrate
19.3 Kinetics of enzyme action
Enzymes are catalysts. They enhance the rate of specific chemical reactions that would occur very slowly in absence of them. They do not change equilibrium point of a reaction nor are they used up or permanently changed by the reactions.
It is found that the substrate concentration has a great effect on initial rate of enzyme catalyzed reactions. All enzymes exhibit saturation effect as explained in section 18.5.2.
This led Victor Henri in 1903 to the conclusion that during enzyme catalyzed reactions, enzyme molecules combine with substrate molecules to form a complex. This idea was expanded into a general theory of enzyme action by Leonor Michaelis and Maud Menten in 1913.
19.4 Michaelis-Menten equation
There are two basic reactions involved in formation and breakdown of enzyme-substrate complex. The enzyme E first combines reversibly with substrate S to form an enzyme-substrate complex ES. This reaction is fast and reversible. The complex ES then breaks down in a second reaction to form product P and the enzyme E is regenerated. This reaction is also reversible but slower than the first reaction.
If [Et] represents total enzyme concentration (the sum of free and combined enzyme), [ES] is the concentration of the enzyme-substrate complex, then [Et]-[ES] represents concentration of free or uncombined enzyme. Substrate concentration [S] is far greater than [Et], so the amount of S bound by E at any given time is negligible compared with total concentration of S.
The rate of formation of [ES] in Equation 1 is
Rate of formation =k1 ([Et ] - [ES])[S] --------- Equation 3
Where k1 is the rate constant of forward reaction 1. The rate of formation of ES from E+P by reverse reaction of (2) is very small and can be neglected.
Rate of breakdown of ES is Rate of Breakdown = k-1(-1) [ES]+ k2 [ES] -------------Equation 4
Where k-1 and k2 are the rate constants for reverse of reaction (1) and reaction (2) respectively.
When rate of formation of ES is equal to its rate of breakdown, concentration of ES is constant and reaction is in steady state.
Thus, equating (3) and (4),
k1 ([Et ]- [ES])[S]=k-1(-1) [ES]+ k2 [ES] ---------------- Equation 5
This can be further solved as
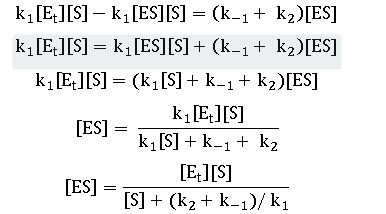
The initial velocity is determined by the rate of breakdown of ES in reaction (2) whose rate constant is k2. Thus we have
v0=k2 [ES]
Thus,
Let, the Michaelis-Menten constant
and , the rate when all available enzyme is present as ES.
Substituting in above equation, we get,
This is the Miscaelis-Menten equation, the rate equation for one-substrate enzyme catalyzed reaction. It is a equation of the quantitative relationship between the initial velocity v0, the maximum velocity Vmax, and the initial substrate concentration, all related through the Michaleis-Menten constant KM.
In the special case when the initial reaction rate is exactly one-half the maximum velocity
when,
Dividing by Vmax and solving for KM,
when,
The Michaelis-Menten equation is basic to all aspects of kinetics of enzyme action. If we know KM and Vmax, we can calculate the reaction rate of an enzyme at any concentration of its substrate.
19.5 Regulation of enzyme activity
Enzymes are catalysts whose activity is regulated by the cell. There are two main reasons for this type of control
(1) All enzyme Action requires energy. A cell has limited store of energy. If an unnecessary enzyme action goes on continuing, the cell will be short of energy and will die.
(2) Some of the products of the enzyme catalyzed reactions may be harmful to the cell. If the concentration of such products increases the cell may die.
Some of the mechanisms which exist for regulation of enzyme activity are as follows:
19.5.1 The simplest method is to produce the enzyme only when it is required. This mechanism is used by bacteria. The bacteria produce enzyme only when a substrate is available to it which it wants to degrade or when a particular product is desired.
19.5.2 Allosterism: Enzymes more than one binding site are called as allosterism enzymes. In such enzymes, one site will be for substrate molecule called as active site. The other site is called as effector binding site. When a molecule (called effector molecule) binds to this site, it changes the shape of active site so that its activity is either increased or decreased. If activity is increased it is called the positive allosterism and if the activity is decreased it is called negative allosterism.
19.5.3 Feedback inhibition: It regulates a chain of reaction involved in synthesis of biological molecules. Consider a sequence of reactions represented below
The starting material A is converted to B by enzyme E1, B is converted to C by Enzyme E2 and so on until final product F is generated. If F is no longer needed, then F will exert negative allosteric effect on enzyme E1 and its activity will be decreased. Thus A will not be converted to B whole chain of relations will stop.
19.5.4 Zymogens: An enzyme in inactive form is called Zymogen or Proenzyme. It is produced away from where it is required. When it reaches its destination, it is converted to active form. E.g. the digestive enzymes pepsin, trypsin and chymotrypsin are very destructive in nature. These enzymes degrade protein to amino acids. If the enzymes are synthesized in active form, the cell will be killed. Thus the cells synthesize inactive forms called pepsinogen, trypsinogen and chymotrypsinnogen which are converted to the active forms in stomach, where they are required for digestion of proteins.
19.5.5 Protein Modification: In this mechanism a covalent group is added or removed from protein molecules of enzyme. This change in enzyme molecule either activates it or turns off its action. Generally, phosphoryl groups are added to or removed from amino acids serine, tyrosine or threonine in protein chain of enzyme.
19.5.6 Inhibition of enzyme activity by pH: Most enzymes are active only within a certain pH range. Making the solutions more basic or acidic causes denaturation of enzyme molecule in which its shape changes and as a result, the activity is lost because the enzyme substrate complex can no longer formed.
References & Further Reading
1. Lehninger A. (1987), “Principles of Biochemistry”. B S Publishers & Distributers, pp. 207-221.
2. Satyanarayana U. & Chakrapani U. (2011), “Biochemistry”. Books and Allied (P) Ltd. pp. 85-103.
3. Denninston (2003), “General, Organic & Biochemistry”. The McGraw-Hill Companies. pp. 612-615.